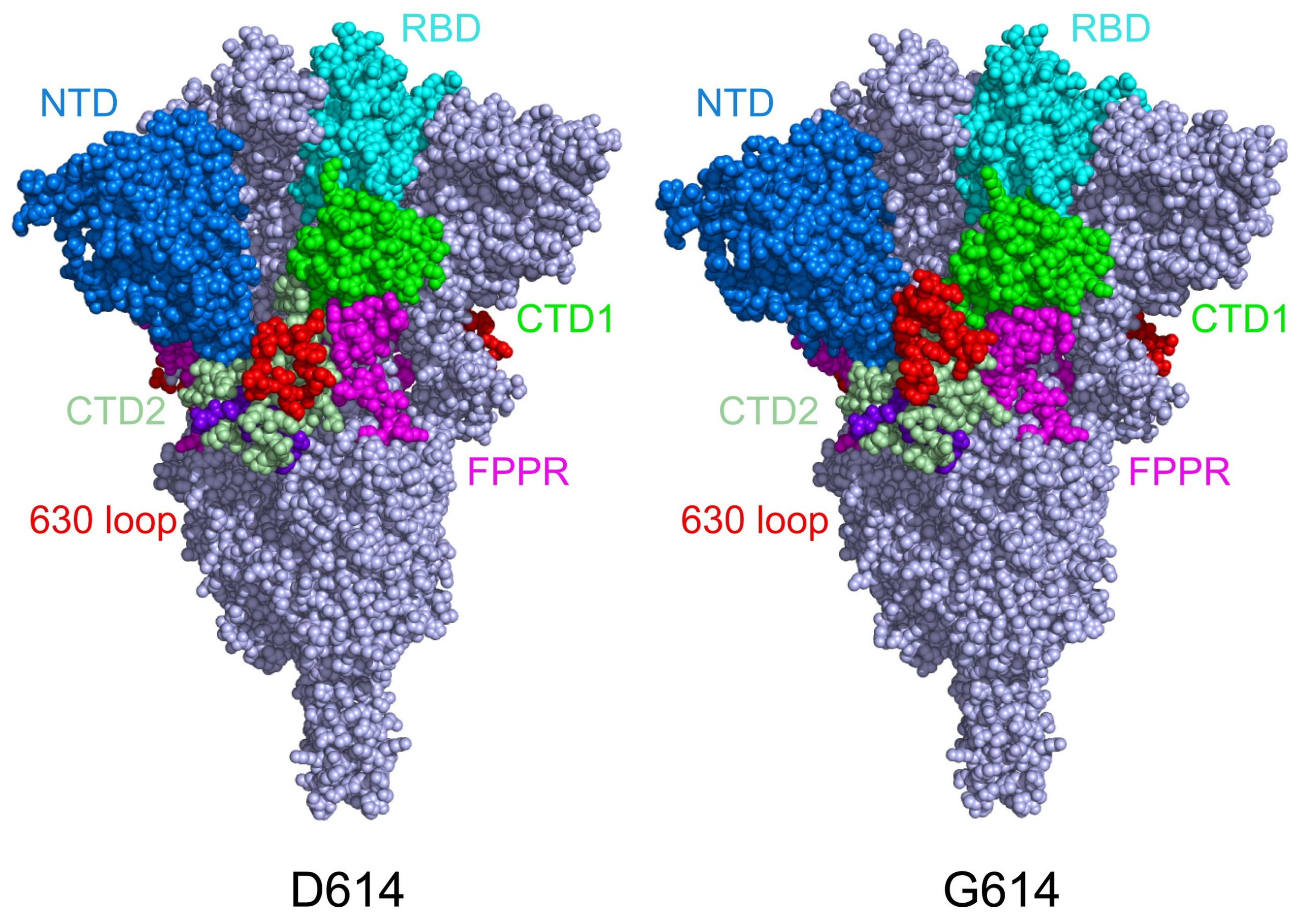
This model shows the structure of the spike protein in its closed configuration, in its original D614 form (left) and its mutant form (G614). In the mutant peak protein, the 630 loop (in red) stabilizes the peak, preventing it from opening prematurely and making SARS-CoV-2 more infectious. Credit: Bing Chen, PhD, Boston Children’s Hospital
The Cryo-EM study shows how structural changes in the G614 variants stabilize the peak.
Rapidly spreading coronavirus variants in the UK, South Africa and Brazil are raising concerns and questions about whether COVID-19 vaccines will protect against them. New work led by Bing Chen, PhD, at Boston Children’s Hospital looked at how the structure of peak coronavirus proteins changes with the D614G mutation – carried by all three variants – and showed why these variants are able to spread more quickly . The team reported their findings in Science on March 16, 2021.
Chen’s team photographed the peaks with cryoelectronic microscopy (cryo-EM), which has resolution down to the atomic level. They found that the D614G mutation (substitution of a single amino acid acid “Letter” in the genetic code for the peak protein) makes the peak more stable compared to the original SARS-CoV-2 virus. As a result, more functional spikes are available to bind to the ACE2 receptors in our cells, making the virus more infectious.
Preventing the change in the shape of the peaks
In the original coronavirus, the peak proteins bound to the ACE2 receptor and changed dramatically in shape, folding over themselves. This allowed the virus to fuse its membrane with the membranes of our own cells and enter. However, as Chen and colleagues reported in July 2020, the peaks sometimes change prematurely and disintegrate before the virus can bind to cells. Although this slowed down the virus, the change in shape also made it more difficult for our immune system to contain it.
“Since the original spike protein dissociated, it was not good enough to induce a strong neutralizing antibody response,” says Chen.
When Chen and his colleagues photographed the peak mutant protein, they found that the D614G mutation stabilizes the peak by blocking premature shape change. Interestingly, the mutation also causes the peaks to bind more weakly to the ACE receptor, but the fact that the peaks are less likely to break down prematurely makes the virus in general more infectious.
“Let’s say the original virus has 100 spikes,” explains Chen. “Because of the instability of the shape, you can only have 50% of them functional. In the G614 variants, you can have 90 percent that are functional, so even though they don’t bond as well, the chances are more likely that you will get an infection. “
Chen proposes that the redesigned vaccines incorporate the code for this peak mutant protein. The more stable peak format should make any peak-based vaccine (such as Moderna, Pfizer and Johnson & Johnson vaccines) more likely to produce protective neutralizing antibodies, he says.
Future direction: a drug to block coronavirus entry
Chen and his colleagues are applying even more structural biology to better understand how SARS-CoV-2 binds to the ACE2 receptor, with a view to therapy to block the virus from entering our cells.
In January, the team showed at Nature Structural & Molecular Biology that a structurally engineered “decoy” ACE2 protein binds to the virus 200 times more strongly than the body’s own ACE2. The bait potently inhibited the virus in cell culture, suggesting that it could be an anti-COVID-19 treatment. Chen is now planning to advance this research on animal models.
Chen is a senior investigator for the newspaper in Science. Jun Zhang and Yongfei Cai, of the Children’s Division of Molecular Medicine in Boston, co-authored. Co-authors were Tianshu Xiao, Hanqin Peng, Sophia Rits-Volloch and Piotr Sliz of Boston Children’s; Jianming Lu of Codex BioSolutions, Inc., Sarah Sterling and Richard Walsh Jr. of the Harvard Cryo-EM Center for Structural Biology (Harvard Medical School); and Haisun Zhu, Alec Woosley and Wei Yang of the Institute for Protein Innovation (Harvard Institutes of Medicine). The work was funded by the National Institutes of Health (AI147884, AI147884-01A1S1, AI141002, AI127193), a COVID-19 award from MassCPR and Emergent Ventures.